nbdev2 - first steps
by Jeremy Howard and Hamel Hussain
- Support
- Walkthrough
- Gitlab integration
- nbdev v2.3
- Open questions
fastai has just released nbdev2.
This is a complete rewrite with quarto. I like how they displayed features in that card
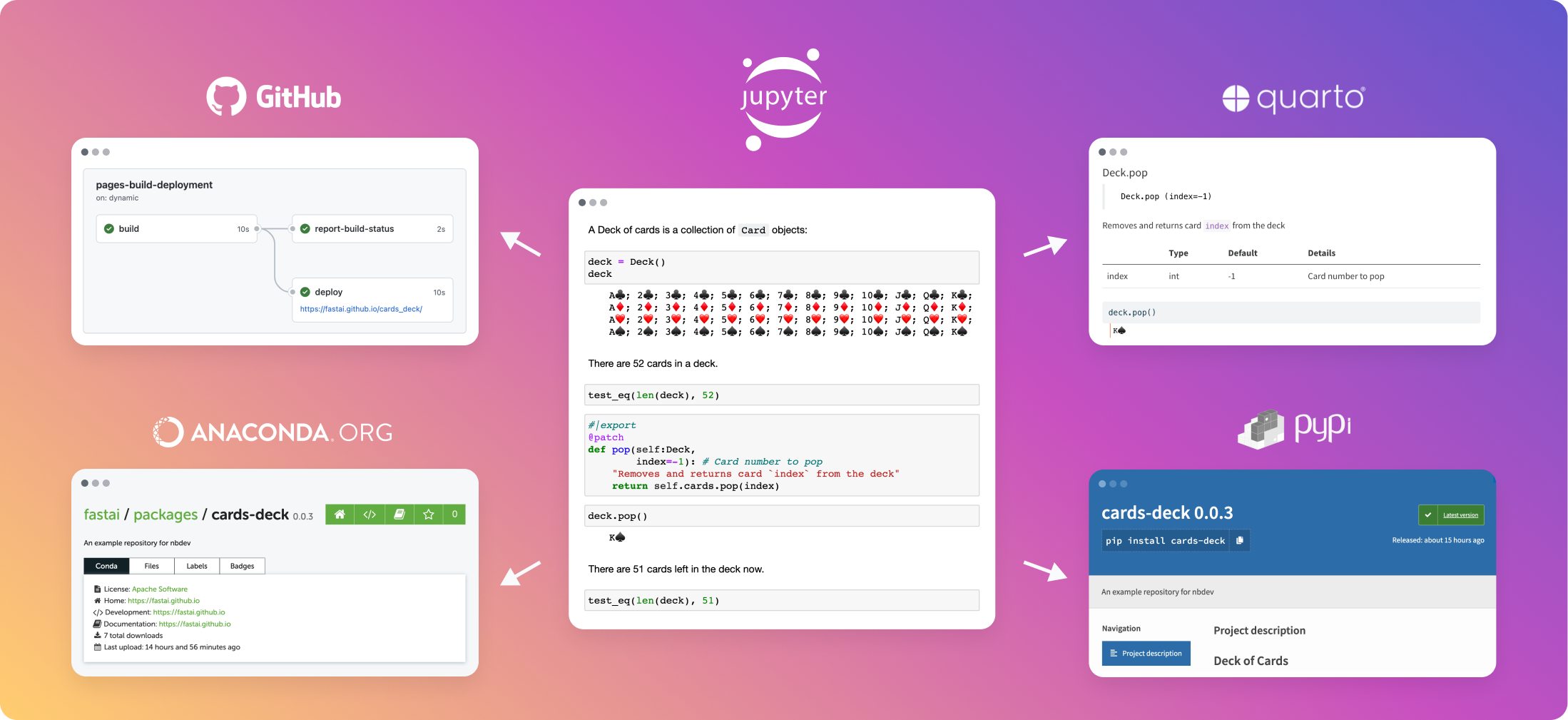
There is a nbdev section in fastai forum.
There is a channel #nbdev-help at fastai discord.
And issues page in github fastai/nbdev repo.
There is a 90 min video: nbdev tutorial -- zero to published project in 90 minutes
I follow here this tutorial.
Here are the big steps:
- create a new project with github: dataset_tools. Give a description it will be reused by nbdev
- create a local conda env
dataset_toolswith what is required to develop this library
!cat /home/guillaume/_conda_env/dataset_tools.txt
import sys
!{sys.prefix}/bin/pip list|grep nbdev
- clone repo
dataset_toolsand turn it into a nbdev repo
git clone git@github.com:castorfou/dataset_tools.git
conda activate dataset_tools
cd dataset_tools
- nbdev can be used from here. For example
nbdev_helpto display all nbdev_ commands and what it does. And more detail can be got with-h:nbdev_new -h
!{sys.prefix}/bin/nbdev_help
-
nbdev_new. It is creating the structure and files such as settings.ini. - from base environment we can start
jupyter notebook. It is advised to install nb_extensions (pip install jupyter_contrib_nbextensions), and activate TOC2. Open00_core.ipynbwithdataset_toolskernel. Rename00_core.ipynb-->00_container.ipynb
Jeremy explains then what are #| used by quarto and nbdev.
And for example #| hide will allow to be executed but hide in your documentation.
Actually from a single notebook, you have 3 usages:
- the notebook by itself - all cells are executed, whatever are the prefix
#|that you display on cells - the python file - only the cells with
#| exportwill be published in a python file referenced as#| default_exp <name of python file>. A new file is genreated whennbdev_exportis called. - the documentation - all cells are used, except the one started with
#| hide. Seems to be dynamically generated (whennbdev_previewis running?).#| exportare handled specifically: if you have import, nothing is displayed. If you have code, definitions and docstrings are exported, and arguments as well.
There is an easy way to describe arguments of a function.
Just make some indentation with comments such as in
def __init__(self,
cle : str, # la clé du container
dataset : pd.DataFrame = None, # le dataset
colonnes_a_masquer : list = [], # les colonnes à masquer
colonnes_a_conserver : list = [] # les colonnes qui ne seront pas transformées
):
and we can directly see the effect of it by calling show_doc (show_doc(Container)). You can even call show_doc on code not written with nbdev, or not even written by you.
There are some basic testing functionalty available by importing fastcore. from fastcore.test import *
With test_eq very closed to assert and test_ne closed to assert not
This is convenient to integrate all the unit tests that way. When you will export by running Restart & Run All, if an error is met, export won't be done.
And one can run nbdev_test from the command line.
Just by adding this import
from fastcore.utils import *
one can use
@patch
def new_method(self:myclass):
pass
from command line, one can run nbdev_export
or directly from jupyter, for example will be executing Restart & Run All
#| hide
import nbdev; nbdev.nbdev_export()
And we can install it to be used directly by running pip install -e .
It means that you can now import your project with
from dataset_tools.container import *
When it will be published (pypi or conda), it will be installable by calling
pip install dataset-tools
or
conda install -c fastai dataset-tools
NB: see how _ has been turned into -, and for that to happen we have to update lib_name and lib_path in settings.ini by replacing _with -
NB2: it is still confusing for me. It looks like modifying lib_path is not a good optiom.
Here it is a good idea to give overview about how to use it.
By importing your library and start using it.
And it will be exported as the homepage of your lib.
Just have to decide what should land in index and what should land in module page.
Just run it from command line
nbdev_preview
and it is accessible from http://localhost:3000.
This is a quarto webserver. The 1st time you launch it it will install quarto for you. On ubuntu this is a standard package so it will be updated regularly.
from getpass import getpass
!echo {getpass()} | sudo -S apt-cache show quarto
As mentionned earlier, one can run nbdev_test to execute all tests in all notebooks.
If it fails, Hamel has shared his dev workflow. He runs Restart kernel & run All, and use %debug magic command to enter debug mode.
You then have access to all ipdb commands such as h for help, p var to print content of var, w for stacktraces
%debug
For a reason it is asked not to mix cells with imports and code.
I am not sure what is the core reason for that. Something due to show_doc or doc generation?
During my tests, I have seen something complaining about it after running nbdev_export or nbdev_test but cannot reproduce that. Hmmm
Just to remove unnecessary metadata in ipynb files.
Will open an issue, because it fails to run here
(dataset_tools) guillaume@LK06LPF2LTSSL:~/git/dataset_tools$ nbdev_clean
/home/guillaume/miniconda/envs/dataset_tools/lib/python3.9/site-packages/nbdev/clean.py:110: UserWarning: Failed to clean notebook
warn(f'{warn_msg}')
/home/guillaume/miniconda/envs/dataset_tools/lib/python3.9/site-packages/nbdev/clean.py:111: UserWarning: clean_ids
warn(e)
/home/guillaume/miniconda/envs/dataset_tools/lib/python3.9/site-packages/nbdev/clean.py:111: UserWarning: clean_ids
warn(e)
Not a bad thing to run all these stuff
nbdev_clean
git diff
git status
git add -A
nbdev_export
nbdev_test
nbdev_docs
git commit -am'init version'
git push
Note that for a reason nbdev_clean is failing
/home/guillaume/miniconda/envs/dataset_tools/lib/python3.9/site-packages/nbdev/clean.py:110: UserWarning: Failed to clean notebook
warn(f'{warn_msg}')
/home/guillaume/miniconda/envs/dataset_tools/lib/python3.9/site-packages/nbdev/clean.py:111: UserWarning: clean_ids
warn(e)
/home/guillaume/miniconda/envs/dataset_tools/lib/python3.9/site-packages/nbdev/clean.py:111: UserWarning: clean_ids
warn(e)
And Hamel suggests to add clean_ids = True in settings.ini
nbdev_docsis pushing the content of index.ipynb to README.md
Just modify settings.inito add dependancies (here pandas)
# end of settings.ini
[..]
### Optional ###
requirements = fastcore pandas
# dev_requirements =
# console_scripts =
clean_ids = True
Et voila!, doc is available at https://castorfou.github.io/dataset_tools/ and you can push that address to your repo settings
This is done by calling nbdev_pypior nbdev_conda.
And it is modifying settings.ini to increment version number. (very much as nbdev_bump_version does)
There are other commands such as nbdev_release_xxx the seems to do quite the same for git.
because this is the platform we use at Michelin, I will need to make it work with our internal gitlab instance.
There is on-going work to make it happen:
- from Hamel Husain - enhancement request Support gitlab
- and from fastai community in forum: Nbdev and Gitlab (source links), Example: nbdev on Gitlab
I have published this version to nbdev forum
Project name : nbdev_gitlab
Project URL : https://gitlab.michelin.com janus nbdev_gitlab
Project description : This is the smallest project to make nbdev working with gitlab
Create project
conda activate dataset_tools
cd ~/git
git clone git@gitlab.michelin.com:janus/nbdev_gitlab.git
export SSL_CERT_FILE='/home/guillaume/miniconda/envs/dataset_tools/lib/python3.9/site-packages/certifi/cacert.pem'
nbdev_new
/home/guillaume/miniconda/envs/dataset_tools/lib/python3.9/site-packages/ghapi/core.py:99: UserWarning: Neither GITHUB_TOKEN nor GITHUB_JWT_TOKEN found: running as unauthenticated
else: warn('Neither GITHUB_TOKEN nor GITHUB_JWT_TOKEN found: running as unauthenticated')
Could not access repo: janus/nbdev_gitlab to find your default branch - `main` assumed.
Edit `settings.ini` if this is incorrect.
In the future, you can allow nbdev to see private repos by setting the environment variable GITHUB_TOKEN as described here:
https://nbdev.fast.ai/cli.html#Using-nbdev_new-with-private-repos
repo = nbdev_gitlab # Automatically inferred from git
user = janus # Automatically inferred from git
author = guillaume # Automatically inferred from git
author_email = guillaume.ramelet@michelin.com # Automatically inferred from git
# Please enter a value for description
description = This is the smallest project to make nbdev working with gitlab
settings.ini created.
/home/guillaume/miniconda/envs/dataset_tools/lib/python3.9/site-packages/ghapi/core.py:99: UserWarning: Neither GITHUB_TOKEN nor GITHUB_JWT_TOKEN found: running as unauthenticated
else: warn('Neither GITHUB_TOKEN nor GITHUB_JWT_TOKEN found: running as unauthenticated')
pandoc -o README.md
to: gfm+footnotes+tex_math_dollars-yaml_metadata_block
standalone: true
default-image-extension: png
metadata
description: This is the smallest project to make nbdev working with gitlab
title: nbdev_gitlab
Output created: _docs/README.md
!ls -l ~/git/nbdev_gitlab
- set company_name = michelin
- set doc_path = public
- set branch = main instead of master
- doc_host = https://%(user)s.pages.gitlab.%(company_name)s.com/
- git_url = https://gitlab.%(company_name)s.com/%(user)s/%(repo)s
- doc_baseurl = /%(repo)s
nothing to be done with nbdev > v2.3.3
With gitlab you have a nice editor to edit pipelines (CI lint)
One way to debug is to insert sleep xx and then click debug.
You then have access to your docker image.
default:
image: 'docker.artifactory.michelin.com/michelin/hub/ubuntu20.04:bib-1.1'
tags:
- k8s
interruptible: true
retry:
max: 2
when:
- runner_system_failure
- stuck_or_timeout_failure
# Functions that should be executed before the build script is run
before_script:
- apt -y install wget
- wget "https://github.com/quarto-dev/quarto-cli/releases/download/v1.1.189/quarto-1.1.189-linux-amd64.deb"
- dpkg -i quarto-1.1.189-linux-amd64.deb
- apt -y install python3-pip
- wget --no-check-certificate --content-disposition -O - https://raw.githubusercontent.com/castorfou/guillaume_blog/master/files/setup_wsl_08_pip.sh | bash
- pip3 install nbdev
- nbdev_install
stages:
- test
- build_doc
- build
- deploy_artifactory
tests:
stage: test
script:
- nbdev_test
pages:
stage: build_doc
script:
- nbdev_docs
artifacts:
paths:
# The folder that contains the files to be exposed at the Page URL
- public
rules:
# This ensures that only pushes to the default branch will trigger
# a pages deploy
- if: $CI_COMMIT_REF_NAME == $CI_DEFAULT_BRANCH
wheel:
stage: build
script:
- mkdir -p public
- echo "Build wheel with python version `python3 --version`:"
- pip install -U setuptools wheel pydnx_packaging
- pip install -e .
- python3 setup.py bdist_wheel
- mkdir -p packages && mv dist/* packages/
artifacts:
when: always
paths:
- packages/
publish:
stage: deploy_artifactory
dependencies:
- wheel
only:
- tags
script:
# create credential config file
- >
if [ -f '.pypirc' ]; then
echo "Information: .pypirc file is not mandatory anymore." && cp .pypirc ~/
else
echo "[distutils]
index-servers = local
[local]
repository: https://artifactory.michelin.com/api/pypi/pypi
username: fm00884
password: <don't even think about it>" > ~/.pypirc
fi
- pip install -U twine
- pip index versions nbdev_gitlab || true
- echo 'If the "twine upload" command below failed with a 403 status code, please check that the version is not already uploaded on artifactory (see versions of nbdev_git above).'
- twine upload --verbose -r local packages/*
nbdev_clean
git diff
git status
git add -A
nbdev_export
nbdev_test
nbdev_docs
git commit -am'init version'
git push
From Settings > General > Badges
create a new entry doc
Link: https://janus.si-pages.michelin.com/nbdev_gitlab/
Badge image URL: https://img.shields.io/badge/-online_documentation-grey.svg
using nbdev_bump_version, nbdev will automatically increase version number
$ nbdev_bump_version
Old version: 0.0.2
New version: 0.0.3
which is modifying 2 files:
$ git diff
diff --git a/nbdev_gitlab/__init__.py b/nbdev_gitlab/__init__.py
--- a/nbdev_gitlab/__init__.py
+++ b/nbdev_gitlab/__init__.py
@@ -1 +1 @@
-__version__ = "0.0.2"
+__version__ = "0.0.3"
diff --git a/settings.ini b/settings.ini
index 1e8adc9..1da37f1 100644
--- a/settings.ini
+++ b/settings.ini
@@ -1,7 +1,7 @@
[DEFAULT]
repo = nbdev_gitlab
lib_name = nbdev_gitlab
-version = 0.0.2
+version = 0.0.3
then one can tag 0.0.3 and push it
git add -A
git commit -am'test tag from git locally'
git tag -a 0.0.3 -m "test tag with git locally"
git push origin 0.0.3 #recommanded way: $git push origin <tag_name>
would be nice to automatically get tag name from settings.ini
New release 2.3 made today (Sept-14)
https://forums.fast.ai/t/upcoming-changes-in-v2-3-edit-now-released/98905
To migrate:
pip install -U nbdev
rm _quarto.yml
nbdev_new
# reintegrate entries in settings.ini that could have been changed such as requirements, dev_requirements, clean_ids
Solution: update fastcore to version > 1.2.5
